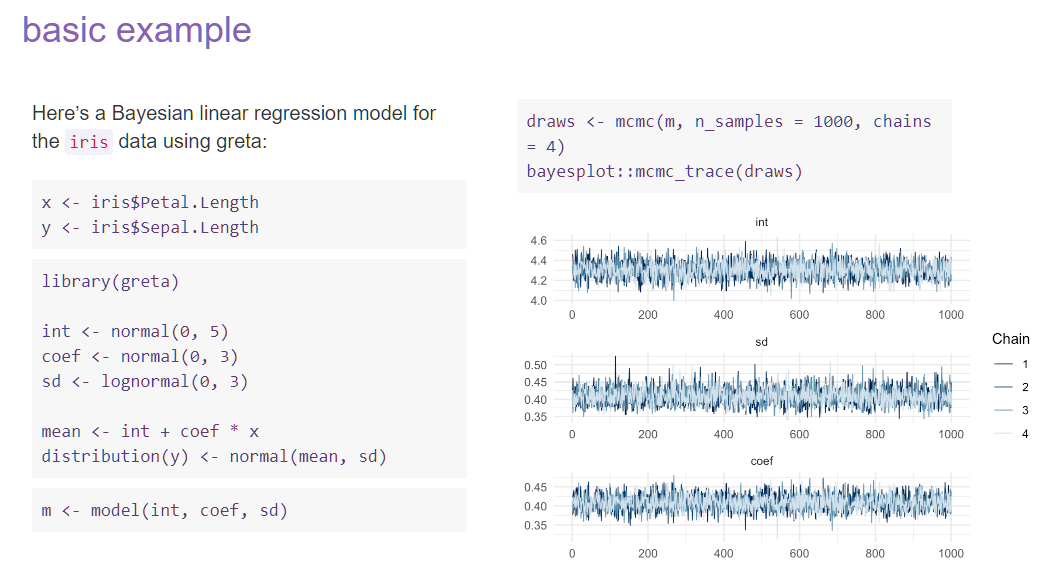
class: center, middle, inverse, title-slide # Bayesian Modeling ## with R2jags ### Zhi Yang ### 2019-04-08 --- # A mini GO~~T~~B family tree 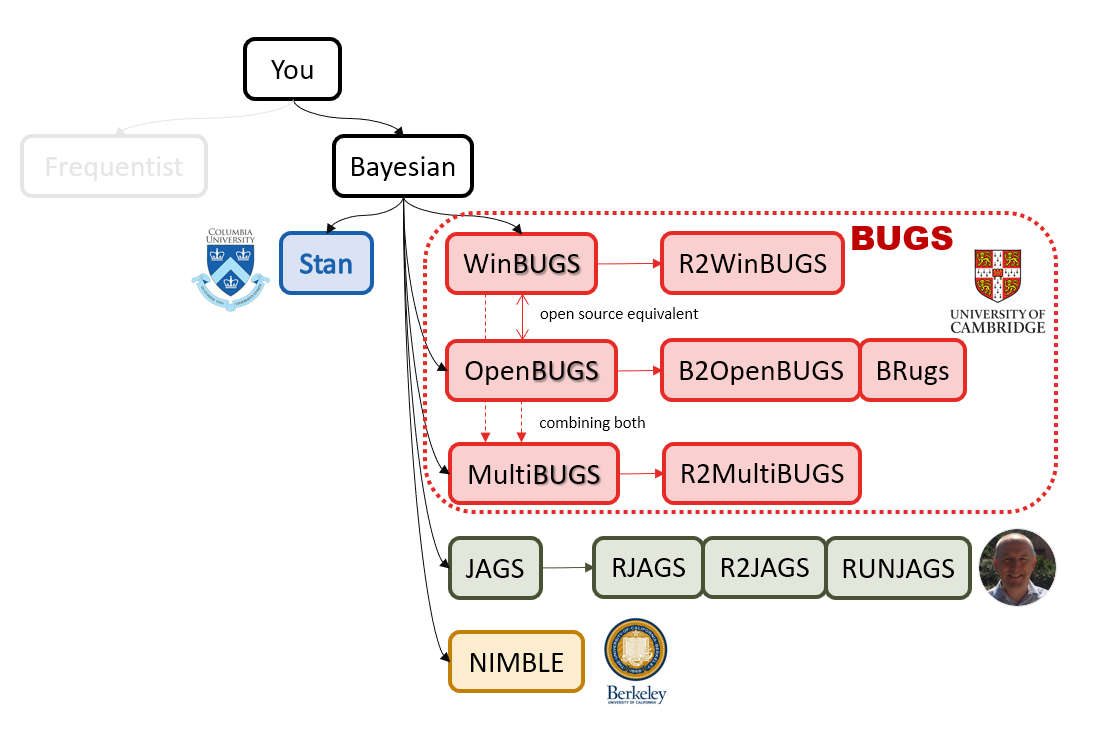 --- # What'll be in this talk #### Some history about tools for Bayesian modeling #### Bayes' theorem, jargons and Shiny demo (prior, posterior ...) #### A walkthrough of using `R2jags` (meetup pizza example) #### More examples (linear regression) --- class: center, middle # .red[Why Bayesian?] -- ### You learn from the data -- ### You know the uncertainty of the estimated parameters .left[.footnote[[My Journey From Frequentist to Bayesian Statistics](http://www.fharrell.com/post/journey/)]] --- class: center, middle #.red[JAGS] -- ###stands for #.red[Just Another Gibbs Sampler] ### developed by Martyn Plummer <img src="imgs/mp.jpg" width="20%"> --- class: center, middle #.red[Why JAGS?] ###why not WinBUGS 🐞 or Stan? --- #JAGS vs. WinBUGS ### WinBUGS is historically very important with slow development -- ### JAGS has very similar syntax to WinBUGS and it allows cross-platform -- ### Great interface to **.blue[R]**! .small[(rjags, R2jags,runjags)] .footnote[ [When there is WinBUGS already, why is JAGS necessary?](https://www.quora.com/When-there-is-WinBUGS-already-why-is-JAGS-necessary) ] --- .pull-left[ # .red[JAGS] * longer history with tons of resources 📕📝🎓 * easy to learn and run it * need to recompile model * fewer developers * less viz tools * various MCMC sampling methods ] .pull-right[ # .blue[Stan] * A newer tool with great team/active community * steeper learning curve * no need to recompile * Syntax check in **.blue[R]** 😱 * ShinyStan (applicable to JAGS) * require fewer iterations to converge ] .center[### Which one's faster? Inconclusive] ### .red[My situation] Stan doesn't support discrete sampling .footnote[ [Stan vs. WinBugs: A search for informed opinions](https://www.reddit.com/r/statistics/comments/5on87q/stan_vs_winbugs_a_search_for_informed_opinions/) ] --- #.red[Why me?] ### I wrote a part of my dissertation with **JAGS** ### I've repeatedly read through **JAGS**, WinBUGS manual and numerous journals ### I've four years' experiences with searching **JAGS** examples in Google and GitHub --- # Installation ### Install **R** ### Install **RStudio** ### Install the **JAGS** from https://sourceforge.net/projects/mcmc-jags/ ### Install the **R2jags** package from [CRAN](https://cran.r-project.org/web/packages/R2jags/index.html): ```r install.packages("R2jags") library(R2jags) ``` --- #30 seconds on Bayes' theorem .pull-left[ 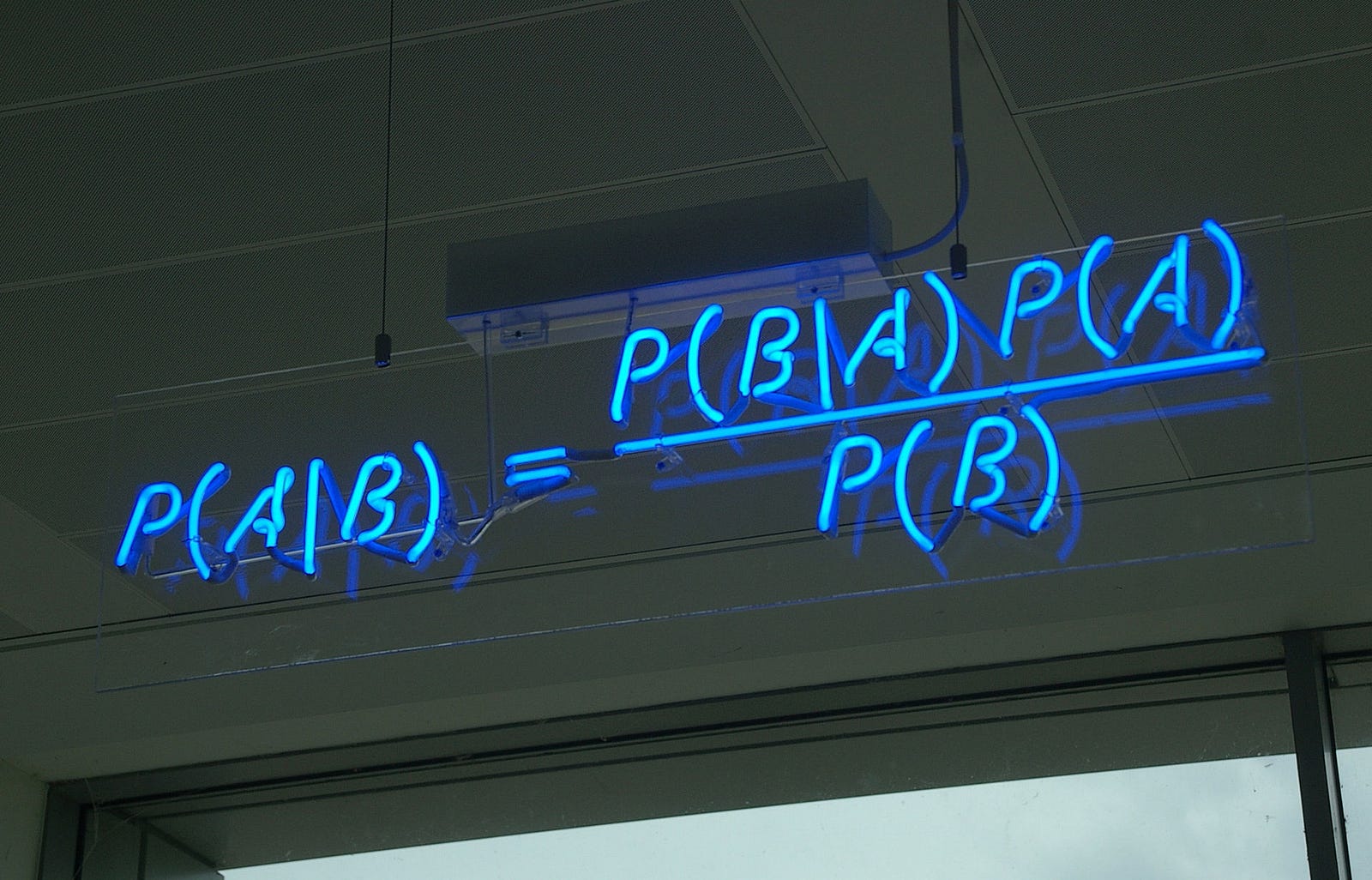 ] .pull-right[ Changing our beliefs based on evidences `$$Posterior = \frac{Likelihood \times Prior}{Evidence}$$` ] ### Prior: strong vs. weak ? <br> ### Data: insufficient vs. sufficient ? .footnote[[Nat Napoletano:Bayes' Theorem for Everyone](https://www.youtube.com/watch?v=XR1zovKxilw)] --- # Shiny Demo The focus of the analysis presented here is on accurately estimating the mean of IQ using simulated data. [FBI: First Bayesian Inference](https://www.rensvandeschoot.com/fbi-the-app/) <img src="imgs/shiny.png" width="90%"> .footnote[https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4158865/] --- # Strong prior vs. weak prior 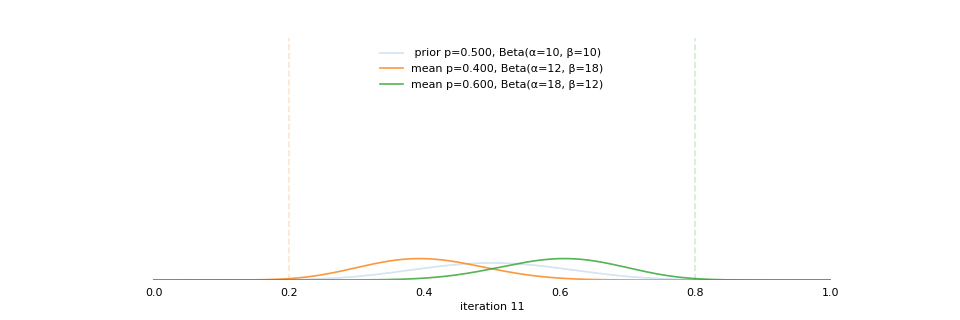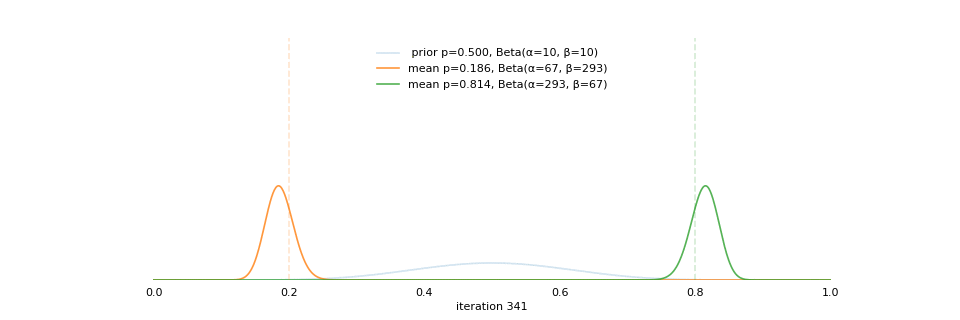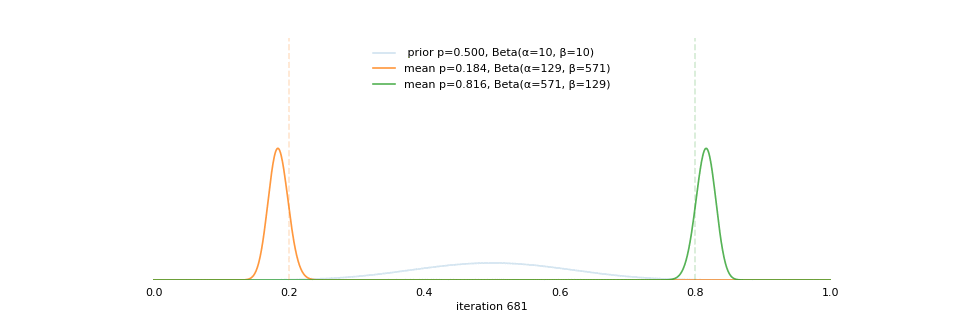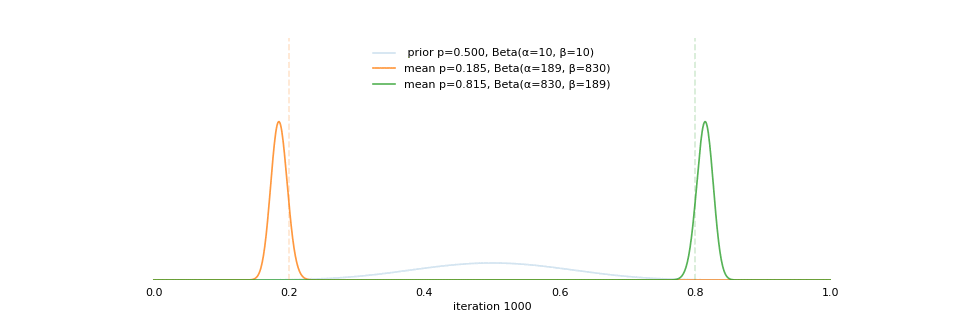  .footnote[https://towardsdatascience.com/visualizing-beta-distribution-7391c18031f1] --- # Let's get started! We hold meetings every month and post it on meetup.com, which tells us how many people've signed up, `\(n\)`. But not everyone show up (i.e., traffic). So, we'd like to know what percentage of people, `\(\theta\)`, will show up to order enough pizzas. .center[ `\(Y_i|\theta \sim Binom(n_i, \theta)\)` `\(\theta \sim Beta(a, b)\)` ] Weak prior: .center[ `\(Beta(1, 1) \leftrightarrow Unif(0, 1)\)` ] ```r Y <- c(77, 62, 30) N <- c(39, 29, 16) ``` --- # Write the model But as you know, someone (@lukanegoita) said <center><img src="imgs/meme.png" width="60%"> --- # Write the model .center[ `\(Y_i|\theta \sim Binom(n_i, \theta)\)` `\(\theta \sim Beta(a, b)\)` ] .pull-left[ In .blue[R] ```r model{ for(i in 1:3){ Y[i] <- rbinom(1, N[i], theta) } theta <- rbeta(1, 1, 1) } ``` ] .pull-right[ In .red[JAGS] ```r model{ for(i in 1:3){ Y[i] ~ dbinom(theta, N[i]) } theta ~ dbeta(1, 1) } ``` ] - A variable should appear **only once** on the left hand side - A variable may appear multiple times on the right hand side - variables that only appear on the right must be supplied as **data** --- # Tips on syntax #### No need for variable declaration #### No need for semi-colons at end of statements #### use d`dist` instead of r`dist` for random sampling #### dnorm(mean, **precision**) .red[not] dnorm(mean, **sd**) #### It allows matrix and array #### It is .red[illegal] to offer definitions of the variable twice --- # Write the model ```r suppressMessages(library(R2jags)) model_string <-" model{ # Likelihood for(i in 1:3){ Y[i] ~ dbinom(theta, N[i]) } # Prior theta ~ dbeta(1, 1) } " # Data N <- c(77, 62, 30) Y <- c(39, 29, 16) # specify parameters to be monitered params <- c("theta") ``` --- # rjags vs. runjags vs. R2jags #### `R2jags` and `runjags` are just wrappers of rjags #### `runjags` makes it easy to run chains in parallel on multiple processes #### `R2jags` allows running until converged and paralleling MCMC chains #### Otherwise, they all have very similar syntax and pre-processing steps --- # Choosing `R2jags` ```r output <- jags(model.file = textConnection(model_string), data = list(Y = Y, N = N), parameters.to.save = params, n.chains = 2, n.iter = 2000) ``` ``` ## Compiling model graph ## Resolving undeclared variables ## Allocating nodes ## Graph information: ## Observed stochastic nodes: 3 ## Unobserved stochastic nodes: 1 ## Total graph size: 9 ## ## Initializing model ## ## | | | 0% | |** | 4% | |**** | 8% | |****** | 12% | |******** | 16% | |********** | 20% | |************ | 24% | |************** | 28% | |**************** | 32% | |****************** | 36% | |******************** | 40% | |********************** | 44% | |************************ | 48% | |************************** | 52% | |**************************** | 56% | |****************************** | 60% | |******************************** | 64% | |********************************** | 68% | |************************************ | 72% | |************************************** | 76% | |**************************************** | 80% | |****************************************** | 84% | |******************************************** | 88% | |********************************************** | 92% | |************************************************ | 96% | |**************************************************| 100% ``` ``` ## Compiling model graph ## Resolving undeclared variables ## Allocating nodes ## Graph information: ## Observed stochastic nodes: 3 ## Unobserved stochastic nodes: 1 ## Total graph size: 9 ## ## Initializing model ## ## | | | 0% | |** | 4% | |**** | 8% | |****** | 12% | |******** | 16% | |********** | 20% | |************ | 24% | |************** | 28% | |**************** | 32% | |****************** | 36% | |******************** | 40% | |********************** | 44% | |************************ | 48% | |************************** | 52% | |**************************** | 56% | |****************************** | 60% | |******************************** | 64% | |********************************** | 68% | |************************************ | 72% | |************************************** | 76% | |**************************************** | 80% | |****************************************** | 84% | |******************************************** | 88% | |********************************************** | 92% | |************************************************ | 96% | |**************************************************| 100% ``` --- # Visualizing output ```r plot(as.mcmc(output)) ``` <!-- --> --- # Visualizing output ```r library(mcmcplots) mcmcplot(as.mcmc(output),c("theta")) denplot(as.mcmc(output),c("theta")) ``` 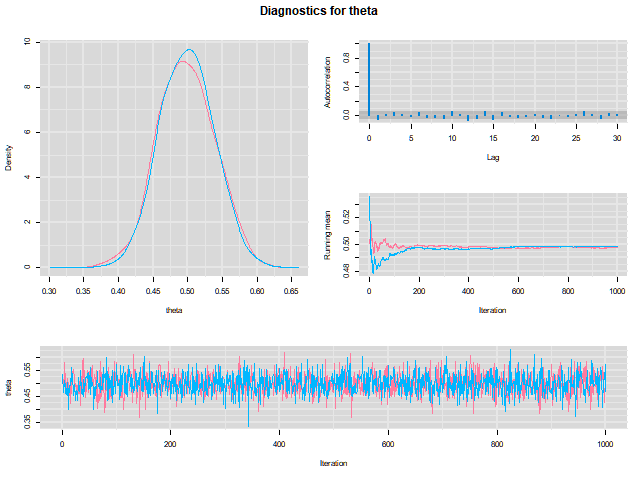 --- # Model diagnostic So it might take a longer time for run <center><img src="imgs/meme2.png" width="60%"> --- ## Model diagnostic and convergence ```r output$BUGSoutput$summary ``` ``` ## mean sd 2.5% 25% 50% 75% ## deviance 14.6685495 1.42564249 13.6497484 13.7569065 14.104791 15.0374241 ## theta 0.4962242 0.03867905 0.4226714 0.4693877 0.496706 0.5209497 ## 97.5% Rhat n.eff ## deviance 18.7817874 1.002871 630 ## theta 0.5723382 1.000970 2000 ``` If it fails to converge, we can do the following, ```r update(output, n.iter = 1000) autojags(output, n.iter = 1000, Rhat = 1.05, n.update = 2) ``` --- # weak prior vs. strong prior ```r output$BUGSoutput$summary ``` ``` ## mean sd 2.5% 25% 50% 75% ## deviance 14.6685495 1.42564249 13.6497484 13.7569065 14.104791 15.0374241 ## theta 0.4962242 0.03867905 0.4226714 0.4693877 0.496706 0.5209497 ## 97.5% Rhat n.eff ## deviance 18.7817874 1.002871 630 ## theta 0.5723382 1.000970 2000 ``` ```r output2$BUGSoutput$summary ``` ``` ## mean sd 2.5% 25% 50% 75% ## deviance 15.2204981 1.7165810 13.6500147 13.9259913 14.6814049 15.9249686 ## theta 0.5346611 0.0298314 0.4752045 0.5146234 0.5355181 0.5547805 ## 97.5% Rhat n.eff ## deviance 19.846200 1.000531 2000 ## theta 0.591993 1.000510 2000 ``` --- # weak prior vs. strong prior .pull-left[ ```r denplot(as.mcmc(output), parms = "theta", xlim = c(0.3, 0.7)) ``` 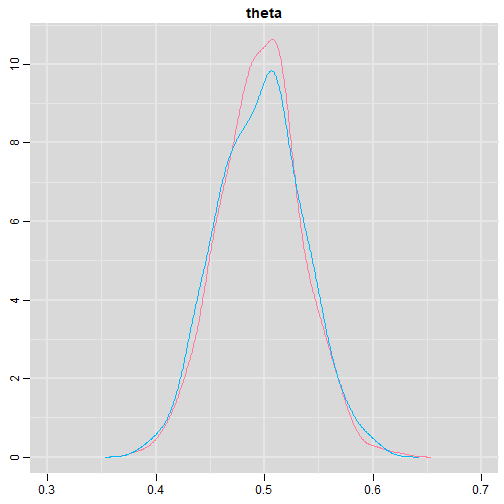<!-- --> ] .pull-right[ ```r denplot(as.mcmc(output2), parms = "theta", xlim = c(0.3, 0.7)) ``` 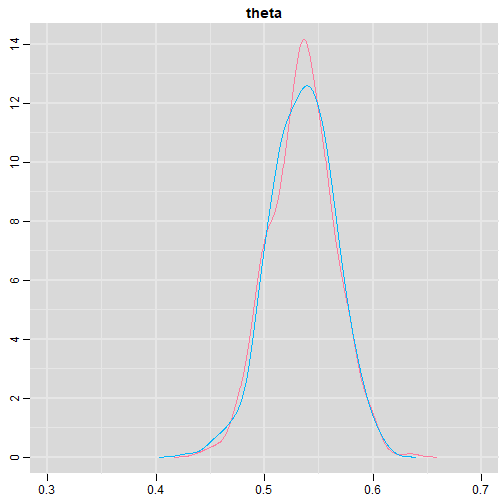<!-- --> ] --- # linear regression .pull-left[ In .red[JAGS] ```r model<-"model{ # Likelihood for(i in 1:N){ y[i] ~ dnorm(mu[i], tau) mu[i] <- beta0 + beta1*x[i] } # Prior # intercept beta0 ~ dnorm(0, 0.0001) # slope beta1 ~ dnorm(0, 0.0001) # error term tau ~ dgamma(0.01, 0.01) sigma <- 1/sqrt(tau) }" ``` In statistics 101 `\(Y_i = b_0 + b_1X_i + \sigma\)` `\(Y \sim N(b_0 + b_1X, \sigma^2)\)` <br> ] .pull-right[ In .blue[R] ```r lm(y~x) ``` ] ``` ## Compiling model graph ## Resolving undeclared variables ## Allocating nodes ## Graph information: ## Observed stochastic nodes: 15 ## Unobserved stochastic nodes: 3 ## Total graph size: 74 ## ## Initializing model ``` --- # Interpretation .pull-left[ In .red[JAGS] ```r lm_output$BUGSoutput$summary[1:2, c(3,7)] ``` ``` ## 2.5% 97.5% ## beta0 -8.853992 52.131691 ## beta1 -16.496277 3.192873 ``` 95% credible interval ] .pull-right[ In .blue[R] ```r confint(lm(y~x)) ``` ``` ## 2.5 % 97.5 % ## (Intercept) -9.32782 51.322335 ## x -16.19329 3.486528 ``` 95% confidence interval ] --- # Resources 📕+💻 .pull-left[ #### If you've got 2hrs: - useR! International R User 2017 Conference JAGS [workshop](https://channel9.msdn.com/Events/useR-international-R-User-conferences/useR-International-R-User-2017-Conference/Introduction-to-Bayesian-inference-with-JAGS) by Martyn Plummer <-- 👨 of **JAGS** ] .pull-right[ #### If you've got 5 mins: [RJAGS: how to get started](https://www.rensvandeschoot.com/tutorials/rjags-how-to-get-started) by Rens van de Schoot, retweeted by Martyn Plummer ] #### <center> If you're a book lover: <center><img src="imgs/book2.png" width=25%> <img src="imgs/book.jpg" width=26%> --- # What about puppies? "perky ears" + "floppy ears" ?= "half-up ears" <img src="imgs/dogs.png"> --- # More ... - [CRAN Task View: Bayesian Inference](https://cran.r-project.org/web/views/Bayesian.html) - ["Selling" Stan](https://discourse.mc-stan.org/t/selling-stan/3693/9) ### Meet greta For those still can't decide which any external tool? - [greta](https://greta-stats.org/index.html) allows you to write models without learning another language like BUGS or Stan - It uses Google TensorFlow (fast!)  --- # greta example